
Home Resources Application notes
Benchmarking LNP vs Electroporation for eGFP RNA and CRISPR-Cas9 Delivery in HSCs
CRISPR–Cas9 gene editing has become a cornerstone technology for therapeutic development, enabling precise genome modification in fields such as immuno-oncology, rare diseases, and regenerative medicine. Its success critically depends on efficient intracellular delivery of Cas9 mRNA and sgRNA, particularly in hematopoietic stems stem (HSCs), which are notoriously difficult to transfect and sensitive to stress.
Abstract
Historically, electroporation has been the standard method for CRISPR delivery in HSCs; however, its high efficiency often comes at the cost of significant cytotoxicity and is limited to exvivo use. In contrast, Lipid nanoparticles (LNPs) have recently emerged as a promising non-viral delivery alternative, offering high biocompatibility, improved cell viability and better translational potential for in vivo use.
This study was conducted in two phases. First, LNPs formulated using the TAMARA microfluidic platform were evaluated for mRNA delivery in HSCs using eGFP-mRNA as a model cargo. Second, LNP-mediated delivery was directly compared with electroporation in a CRISPR knockout assay targeting the β2-microglobulin (B2M) gene.. Transfection efficiency, genome-editing activity, and post-transfection viability were quantified to assess and benchmark both approaches.
The results demonstrated that LNPs enable highly efficient RNA delivery to HSCs with minimal cytotoxicity, achieving gene-editing levels comparable to electroporation. While LNP formulations may require further optimization depending on payload and target cells, they represent a scalable, less invasive, more viable and translationally relevant alternative to electroporation for RNA-based genome editing in hematopoietic models.
An in vitro comparison of mRNA and CRISPR-Cas9 transfection efficiency and genome-editing performance using lipid nanoparticles and electroporation in primary hematopoietic stem cells.
Introduction
CRISPR–Cas9 technology has revolutionized genome engineering, enabling precise and programmable editing for therapeutic applications. Its use is expanding rapidly across fields such as immuno-oncology, hematology, and regenerative medicine. Discovered in the early 2000’s from bacterial adaptive immunity, the CRISPR–Cas9 system delivery has been improved using sgRNA to guide the Cas9 endonuclease to the targeted genomic sites. Later, the convergence of synthetic mRNA and lipid nanoparticle (LNP) technology has offered an elegant solution to reduce off-target activity by yielding transient Cas9 expression. Efficient co-delivery of Cas9 mRNA and sgRNA is thus critical to achieving robust and reproducible gene editing.

Among target cell types, hematopoietic stems stem (HSCs) are of particular interest due to their central role in blood and immune system regeneration, as well as their potential for curative gene therapies. However, these cells are notoriously difficult to transfect because of their quiescent state and fragile membrane integrity, making the choice of delivery method a key determinant of success.
Electroporation has long been the reference technique for introducing RNA or CRISPR reagents into HSPCs, yet it is associated with high levels of cellular stress and loss of viability. Lipid nanoparticles (LNPs), by contrast, have recently emerged as a the gold standard non-viral RNA delivery platform, offering scalability, tunable composition, and compatibility with in vivo and ex vivo applications.
In this study, we first validated eGFP mRNA delivery in HSCs using LNPs to assess baseline transfection efficiency and cell viability at different RNA doses. Building on this, we evaluated the co-delivery of Cas9 mRNA and sgRNA in a CRISPR knockout (KO) model targeting β2-microglobulin (B2M), a protein uniformly expressed on the cell surface and readily quantifiable via flow cytometry, to compare genome-editing performance. LNPs were formulated using the TAMARA microfluidic system to ensure precise, reproducible, and scalable nanoparticle production across R&D scale. The goal of this study is to benchmark both delivery approaches—electroporation and LNP-mediated delivery—by assessing transfection efficiency, editing activity, and post-transfection viability, to help researchers identify the most suitable RNA delivery strategy for gene editing in hematopoietic models.
This work was carried out in partnership with the BIGRes team (B-cell Ig Gene Remodeling Singularities) a joint EFS & Inserm lab led by Michel Cogné in Rennes, France, with significant contributions from Dr. Gregory Noel and supported by Inside Therapeutics.
Results
Overview
Performance evaluations were conducted in two stages. First, an eGFP mRNA model was used to assess dose-dependent RNA delivery in HSC cells using a Moderna-like SM-102 LNP formulation. Second, a proof of concept study was performed using Cas9 mRNA and sgRNA targeting the β2-microglobulin (B2M) locus. Finally, a comparative study with electroporation was performed using this KO model.
For detailed preparation and composition information, see the Materials and Methods section.
1/ Physicochemical characterization of the LNP
LNPs were characterized by NTA following formulation. Both eGFP and Cas9-containing LNPs presented mean sizes near 100 nm, aligning with established standards for efficient intracellular delivery.
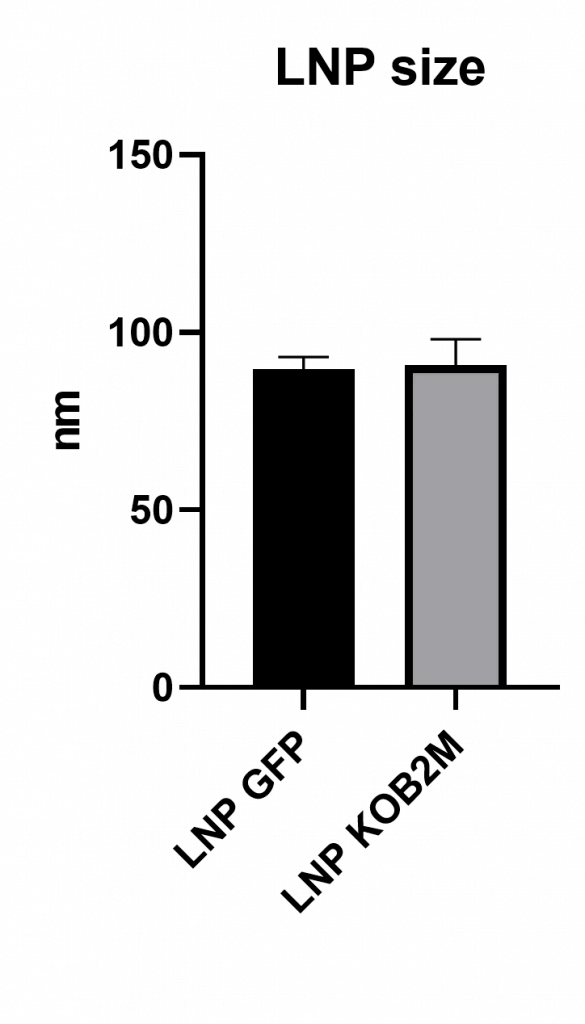
2/ Hematopoietic stems cells transfection
A. eGFP mRNA delivery via LNP in HSCs
In a first place, HSCs were treated with RNA-LNP encapsulating eGFP formulated using the standard Moderna formulation (SM-102) at increasing RNA doses (0.125, 0.25, and 0.5 µg for 200.000 HSC cells).
Transfection results can be observed below, showing the percentage of cells becoming eGFP+.
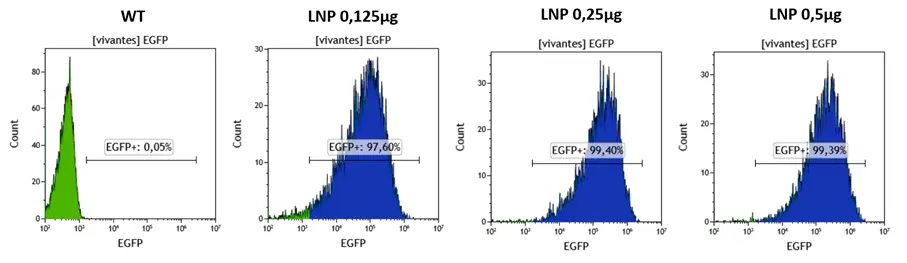
Flow cytometry revealed near-complete transfection across all doses, with eGFP-positive cells approaching 100 percent, showing excellent responses of the HSCs cells.

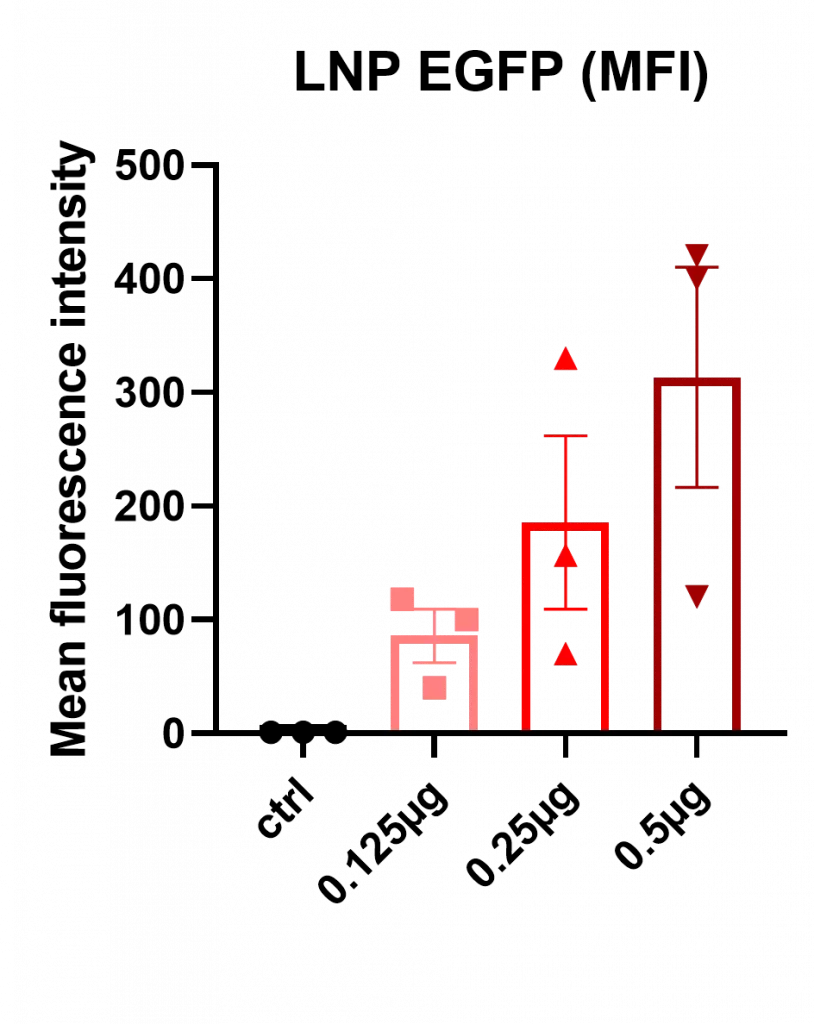
In a second step, Mean Fluorescence Intensity (MFI), used as a proxy for intracellular mRNA levels, increased proportionally with RNA dose, consistent with expected dose-response behavior.
Finally, cell viability remained excellent, with less than 1 percent cell death observed in all conditions.
B. Cas9 mRNA + sgRNA codelivery in HSC via LNP
Next LNPs containing Cas9 mRNA along with guide RNAs targeting an exon of the Beta-2-microglobulin gene were formulated using standard formulation (SM-102) at different RNA doses (0.125, 0.25, 0.5 and 1 µg for 200.000 HSC cells). Since the B2M protein is expressed on the cell surface, the knockout gene can be easily quantified by flow cytometry..
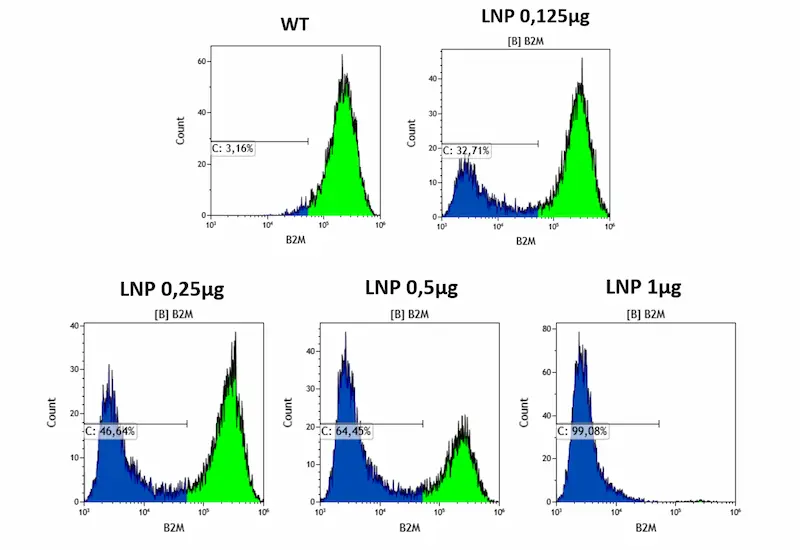
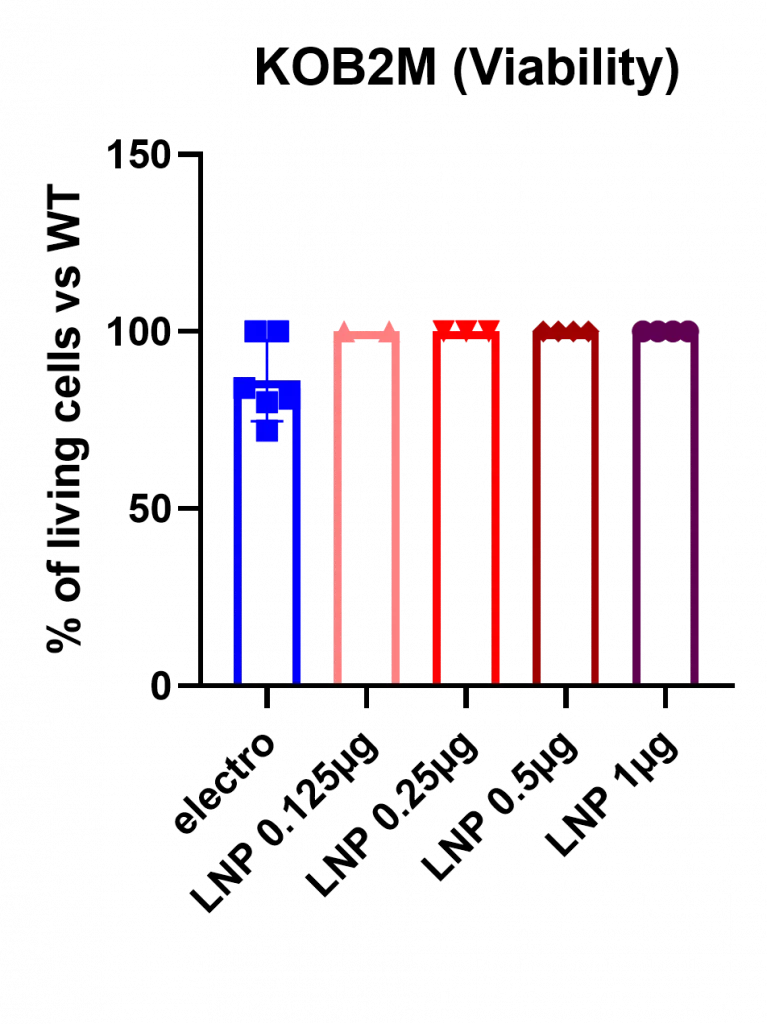
As expected, KO efficiency displayed a clear dose dependency, with the largest dose (1 µg) achieving a near 100% knock-out of the B2M gene. Also, as with eGFP mRNA, viability remained excellent (<1 percent cell death).
C. Comparing Electroporation and LNP for CRISPR in HSC
Finally, we benchmarked LNP-mediated CRISPR delivery against electroporation using identical Cas9/sgRNA components targeting the B2M locus.
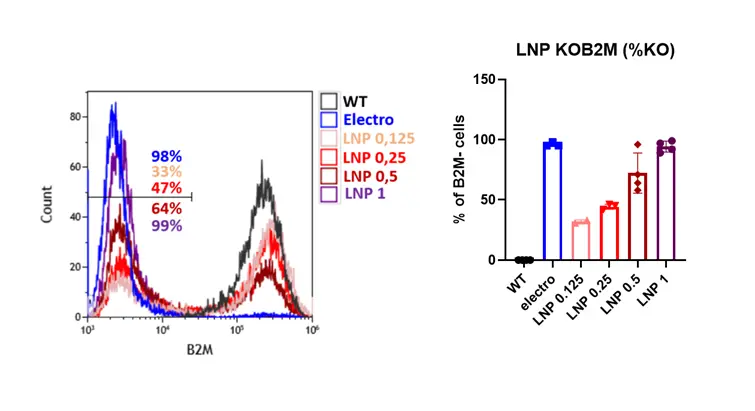
Both approaches achieved similarly high knockout efficiencies, reaching ~100% B2M disruption at their optimal conditions. However, a marked difference was observed in cytotoxicity: electroporation caused substantial cell loss (~20% cell death), whereas LNP-mediated delivery consistently preserved near-complete viability (~99%), as shown in Figure 7. This difference is particularly important for applications where cell recovery is limiting—such as primary HSC editing for transplantation.
In addition to improved viability, LNPs enabled a far more efficient use of CRISPR reagents. Because the two methods relied on different Cas9 formats (mRNA for LNP delivery vs. protein for electroporation), direct comparisons of Cas9 doses were not performed. However, the amount of sgRNA required per million cells showed a striking difference: electroporation required ~3.2 µg of sgRNA per million cells, whereas LNP-mediated delivery achieved comparable editing using only ~0.32 µg per million cells—an order of magnitude reduction. This lower RNA demand significantly decreases reagent costs and enhances scalability, while facilitating future GMP manufacturing and translational applications.
Conclusion & discussions
This study demonstrates that hematopoietic stem cells can be efficiently transfected with both eGFP mRNA and CRISPR reagents using a standard SM-102 lipid nanoparticle formulation. LNPs produced with the TAMARA microfluidic system showed excellent physicochemical properties and consistently delivered high transfection efficiencies, clear dose–response behavior, and near-zero cytotoxicity.
When applied to CRISPR–Cas9 delivery, LNPs achieved knockout levels comparable to electroporation while maintaining almost complete cell viability—an important advantage for workflows involving sensitive primary cells or applications in which cell recovery and functional integrity are critical.
Moreover, LNP-based delivery required an order of magnitude less sgRNA per cell than electroporation, underscoring substantially improved reagent efficiency.
Taken together, these results position LNP-mediated delivery as a gentle, scalable, and more translationally relevant alternative to electroporation for gene-editing workflows in hematopoietic models. Whereas electroporation remains restricted to ex vivo manipulation, LNPs offer the additional advantage of compatibility with in vivo administration, opening avenues toward direct HSC-targeted editing and broader therapeutic strategies that cannot be achieved by electroporation-based approaches.
Despite the strong performance observed here, several opportunities remain to further enhance LNP-mediated CRISPR editing. Next-generation ionizable lipids—such as LP-01, originally developed by Intellia—may further improve endosomal escape and boost editing efficiency. In addition, alternative delivery strategies, such as optimizing co-delivery of Cas9 mRNA and sgRNA, or oppositely evaluating separate delivery timing for the mRNA-coding for Cas9 and sgRNA components, may reveal kinetic advantages that further enhance editing efficiency.. Future studies integrating advanced lipid chemistries, optimized LNP architectures, and targeted delivery approaches may elevate CRISPR editing in HSCs even further, potentially surpassing current electroporation benchmarks while preserving superior viability and translational potential.
Material and methods
RNA-LNP Formulation
RNA-LNP composition
eGFP RNA : eGFP mRNA cleancap
Cas9 mRNA: Codon-optimized Cas9 mRNA
sgRNA: Proprietary design by the BIGRes team Lipid Composition : Moderna like composition including SM-102 ionizable lipid.
Composition can be found here in our LNP starter kits
RNA-LNP formulation systems: TAMARA
TAMARA is an advanced nanoparticle / RNA-LNP formulation system developed by Inside Therapeutics.
This microfluidic-based platform is designed to support the entire R&D process for RNA-LNP medicines and therapies, from the first screening steps to in-vivo testing.
It incorporates a reusable chip with an optimized fluidic design that eliminates dead volumes. Its high-end microfluidic mixing allows for the highest level of encapsulation efficiency as well as control and repeatability of the produced LNP.
For those reasons TAMARA offers a highly efficient and cost-effective solution for RNA-LNP development.

TAMARA offers 2 microfluidic mixing approach within one chip that the user can select at will: an optimized Herringbone mixer and a Baffle mixer. Throughout this entire study, the Herringbone mixing approach was employed.
Formulation protocol
A standard formulation protocol was used for the formulation of RNA-LNP. Formulation parameters used:
- FRR: 1:3
- TFR: 5 mL/min
- Post formulation process: Ultrafiltration using amicon filters
More details on the formulation protocol can be found in the “TAMARA standard protocol” available on Inside Therapeutics’s website.
Characterization
RNA-LNP size
RNA-LNP size measurement was carried out using an NTA (Nanoparticle tracking analysis)
Cell assays
T -1: Transfection Day -1: 200,000 HSCs were seeded in the appropriate well plate, such that the cells were at 70-80% confluent on the end point.
T0: Transfection Day:
· Media was removed from the cells
· mRNA-LNP was added at the specified RNA concentration
· Cells were incubated with the treatment for 24 hours
Flow cytometry
Post transfection, the protein expression, has been assessed by FACS.


Looking to get started or improve your LNP formulation screening?
Reach out to us to discover how we can help!
Other Application notes
Looking for more detailed application notes on nanoparticle formulations? Check out our other resources!
See all Application notes

