
Abstract
This guide outlines modern strategies for RNA-LNP formulation screening, emphasizing the challenges of optimizing critical process parameters (CPPs) and quality attributes (CQAs). It highlights how tools like Design of Experiments (DOE), AI, and especially microfluidic platforms such as TAMARA enable precise, high-throughput, and cost-effective LNP development and LNP screening for RNA therapeutics.
Lipid nanoparticles (LNPs) have become indispensable tools in the development of RNA vaccines and RNA-based therapeutics such as the mRNA LNP Covid 19 vaccines. These nanocarriers play a pivotal role as non-viral delivery vehicles, protecting fragile RNA molecules as they traverse the body and ensure their efficient delivery into target cells and organs. By enabling RNA therapies to achieve their therapeutic potential, LNPs have become a cornerstone of modern medicine opening the door for numerous new gene therapies and vaccine development.
A critical challenge in those novel RNA LNP drug development lies in the screening of LNP to optimize their RNA delivery efficiency while minimizing toxicity and precisely controlling biodistribution to achieve favorable therapeutic outcomes.
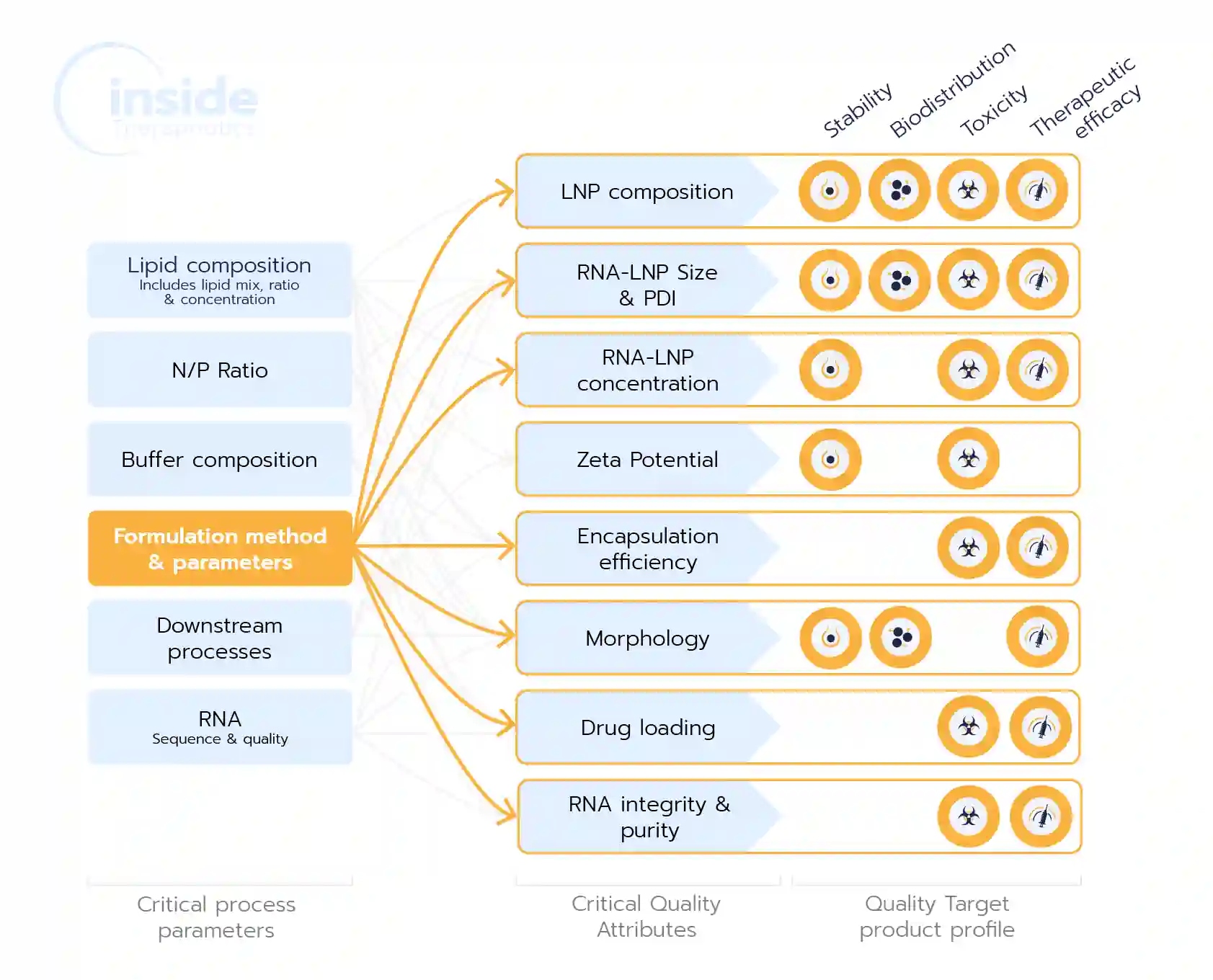
This process depends on fine-tuning the interplay between critical process parameters (CPPs)—such as lipid composition, lipid ratios, buffer conditions, ligands, and formulation process parameters—and their influence on critical quality attributes (CQAs) of the LNP, such as size, polydispersity index (PDI), zeta potential, morphology, and encapsulation efficiency… which in turn directly affect the Quality Target Product Profile (QTPP), such as therapeutic efficacy, toxicity, and biodistribution of the nanoparticles.
A key consideration is that, while general guidelines or “rules of thumb” exist, there is no straightforward or universally defined correlation between CPPs, CQAs, and QTPP. This lack of predictability makes empirical testing the cornerstone of LNP screening and LNP optimization. However, this process is inherently challenging: even when focusing solely on lipid composition, the number of potential combinations—including multiple lipid types, varying ratios, and additional parameters—can quickly lead to thousands or tens of thousands of possibilities. Yet just this simple optimization on lipids already has the potential to yield improvements in mRNA delivery efficiency by at least to 100-fold [1]
To address this immense experimental landscape, sophisticated strategies are needed to streamline the screening process. Adding to this complexity is the high cost of critical materials, such as RNA and novel lipid components, which can limit the feasibility of large-scale testing. Advanced methodologies, such as design of experiments (DOE) and AI-driven optimization, offer promising solutions to identify high-performing candidates while significantly reducing the number of required tests. Furthermore, the formulation process itself must be optimized for high throughput while minimizing RNA and lipid consumption to mitigate costs and accelerate timelines.
In this article, we will explore the most critical aspects of LNP formulation screening. We will begin by examining the key parameters to evaluate and observe, followed by an exploration of strategies to optimize the screening process. By balancing efficiency and cost, these approaches aim to enable the development of superior LNP formulations, accelerating progress and enhancing the quality and effectiveness of RNA therapeutics.
Reminder on LNP Formulation CPPs and CQAs
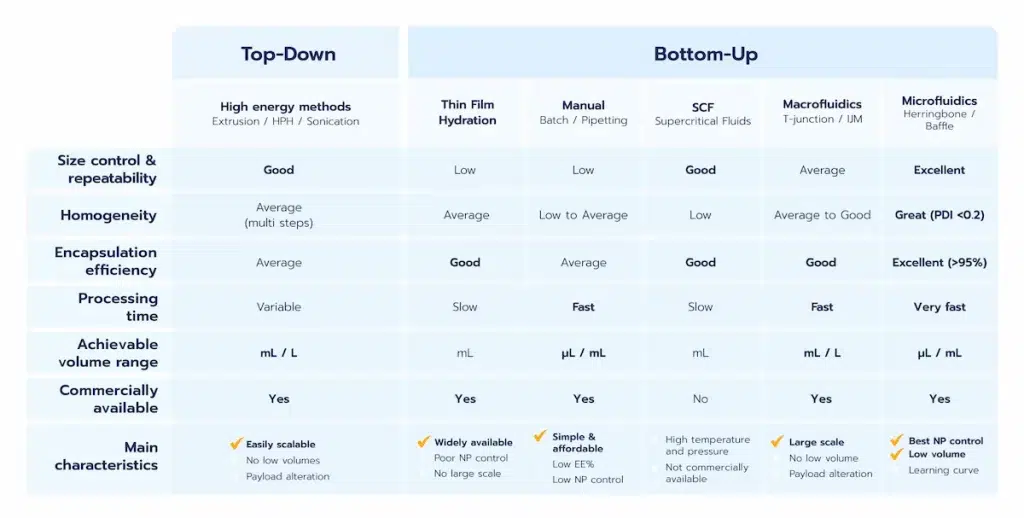
In the formulation of lipid nanoparticles (LNPs), understanding and optimizing critical process parameters (CPPs) and critical quality attributes (CQAs) is essential to ensure the production of safe, effective, and reproducible RNA-based therapeutics. Below, we revisit these parameters, detailing their roles and impacts, and summarize key CQAs in a table for clarity.
Critical Process Parameters (CPPs) of the LNP
1 – Lipid Composition
- Ionizable lipid: The primary component of the LNP. Neutral at physiological pH (~7), it becomes positively charged below its pKa, facilitating RNA and nucleic acid encapsulation, endosomal escape, and reducing toxicity.
- Phospholipid: it stabilizes the LNP structure and enhances encapsulation efficiency.
- Sterol (e.g., cholesterol): Provides rigidity, improves LNP stability, and reduces drug leakage.
- PEG lipid: Found mostly on the surface of the LNP, PEG lipids influence nanoparticle size and enhance circulation time by reducing aggregation and improving the stealth effect.
Overall, the lipid composition affects all nanoparticle characteristics
2 – Lipid Ratios
- The specific ratios of the four lipid components influence the CQAs such as LNP size, encapsulation efficiency, and stability. For instance, when increasing the PEG ratio, nanoparticle size will tend to decrease.
- Typical lipids ratios used in commercially available mRNA vaccines are:
- Pfizer-BioNTech’s BNT162b2: 46.3:9.4:42.7 :1.6 (ionizable lipid, sterol , phospholipid, PEG lipid). [2]
- Moderna’s mRNA-1273 & Alnylam’s Onpattro: 50:10 :28 :1.5 [2]
| Lipid Class | Ionizable Lipid | PEG | Phospholipid | Sterol |
|---|---|---|---|---|
| Lipid Name | ALC-0315 | ALC-0159 (PEG2000) | DSPC | Cholesterol |
| Molar Ratio | 46.3 | 1.6 | 9.4 | 42.7 |
3 – RNA properties
- The characteristics of the RNA/Nucleic acid sequence significantly influence the final properties of the RNA–LNP complex, explaining why a “one size fits all” approach unfeasible.
- The purity of the RNA/Nucleic Acid and presence of dsRNA plays a critical role in determining various RNA–LNP attributes, including encapsulation efficiency and polydispersity index (PDI).
4 – N/P Ratio
- The N/P ratio measures the molar ratio of the amine groups (N) in the ionizable lipid to the phosphate groups (P) in the RNA cargo.
- Typical ranges are 3:1 to 12:1, with Pfizer and Moderna both utilizing a 6:1 ratio. This parameter influences RNA encapsulation, endosomal escape, and toxicity.
5 – Buffer Composition
- Buffers must be carefully chosen to maintain the integrity of both lipids and RNA.
- The pH and ionic strength of the buffer during formulation affect the ionization state of lipids and thus influence LNP size and encapsulation efficiency.
6 – Formulation Method
- LNPs are typically formulated using bottom-up methods where two phases are mixed to trigger nanoparticle self-assembly.
- The mixing technique—such as manual mixing, macrofluidic or microfluidics—significantly affects the LNP formation process, generally by self assembly, and thus all the CQAs such as size, encapsulation efficiency, morphology…
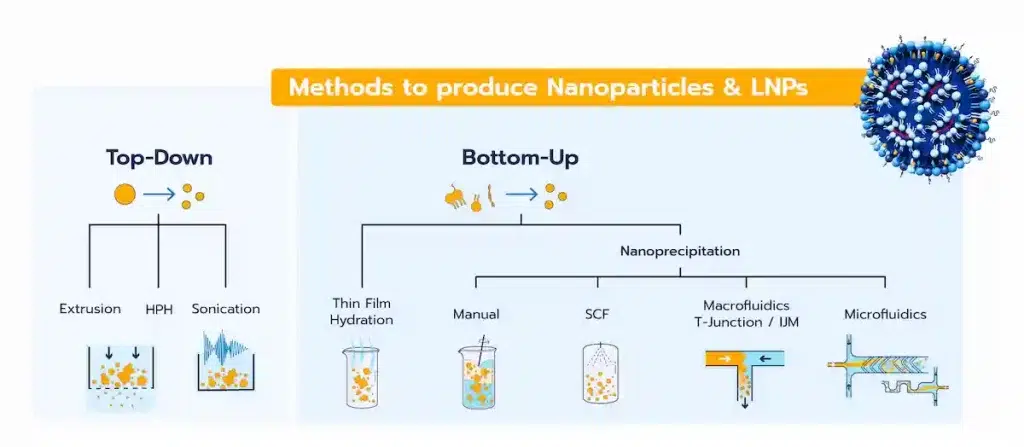
7 – Formulation Parameters
- Depending on the formulation method used, the formulation parameters will impact sthe self-assembly process too.
- Microfluidics, for example, enables precise control of parameters like flow rate ratios (FRR) and total flow rates (TFR), directly impacting particle size and uniformity. For instance, in microfluidics, higher TFRs typically result in smaller LNPs.
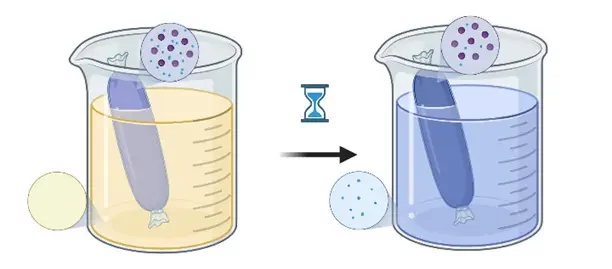
8 – Downstream Processing
- Solvent removal methods such as dialysis, tangential flow filtration (TFF), or ultracentrifugation influence final particle characteristics and stability.
LNP Critical Quality Attributes (CQAs)
The CQAs define the physical and chemical properties of LNPs that directly impact their safety, efficacy, and reproducibility. Even minor deviations can lead to changes in therapeutic performance. Key CQAs are summarized in the table below
| CQA | Desciption | Typical LNP Characterization Method | Impact |
|---|---|---|---|
| LNP Composition | Lipid composing the LNP & ratio of the different lipids | Liquid chromatography | Impacts RNA stability, encapsulation & delivery efficiency, stability, and therapeutic efficacy. |
| Size | Nanoparticle mean diameter. Typical mean nanoparticle size range: 60-120 nm | DLS, NTA | Affects cellular uptake, biodistribution, circulation time, and toxicity. |
| Polydispersity Index | Measures size uniformity; lower PDI indicates higher uniformity (<0.2 is ideal). | DLS, NTA | Ensures consistent therapeutic performance and reduces off-target effects & toxicity. |
| Zeta Potential | Reflects surface charge, influenced by lipid composition. | DLS | Affects LNP stability (low charge) and toxicity (high charge). |
| Encapsulation Efficiency | Percentage of RNA successfully encapsulated (>90% is optimal). | Fluorescence-based assay | Poor encapsulation increases free RNA, raising toxicity and reducing therapeutic efficacy. |
| Morphology | Includes factors like blebs, full/empty ratio, and RNA copy number per LNP. | Cryo-TEM | Influences internalization, delivery efficiency, and stability. |
| Drug Loading | Mass ratio of drug to drug-loaded nanoparticles. | Nano flow cytometers | Therapeutic efficacy and toxicity. |
| RNA Purity | Evaluate the presence of impurities or contaminants in the RNA. | High-performance liquid chromatography | Can trigger an immune response and/or increase toxicity. |
As one can notice, there are therefore multiple interdependencies Between CPPs and CQAs and the final characteristics of the drug product such as;
- Lipid Composition and Ratios: Impacts all process parameters, and thus all QTPPs.
- Formulation Method and Parameters: Affect nearly all the CQAs, and therefore all the parameters of the final drug product.
In the context of screening, researchers should then seek to optimize these CPPs and monitor those CQAs, to develop novel LNP formulations that maximize RNA delivery efficiency, minimize toxicity, and ensure reproducibility.
& Quality attributes of the drug product.
LNP Screening Optimization Approaches
Considering the multitude of critical process parameters (CPPs) to be tested and their interdependencies, the primary goal in LNP screening is to minimize the number of formulations required to identify the optimal working conditions for a candidate. Ideally, this process should lead to a predictive model for future formulations. Two key strategies have emerged in this space: Design of Experiments (DOE) and the increasingly explored role of Artificial Intelligence (AI) in LNP formulation.
Design of Experiments (DOE) in LNP Screening
Design of Experiments (DOE) is a systematic and efficient approach to exploring the relationships between multiple factors and their impact on outcomes, making it particularly well-suited for LNP screening. In the context of LNP development, DOE allows researchers to evaluate the influence of CPPs on critical quality attributes (CQAs) in a structured manner. This method identifies optimal formulations with significantly fewer experiments compared to traditional trial-and-error approaches.
By employing statistical models, DOE enables the simultaneous evaluation of multiple variables, such as lipid composition, lipid ratios, and formulation parameters, as well as their interactions. This approach not only reduces resource consumption but also provides deeper insights into how these factors collectively influence key attributes like encapsulation efficiency, particle size, and biodistribution. As a result, DOE accelerates the identification of robust LNP formulations, decreasing the number of required test formulations while ensuring a balance between efficiency, cost, and therapeutic performance.
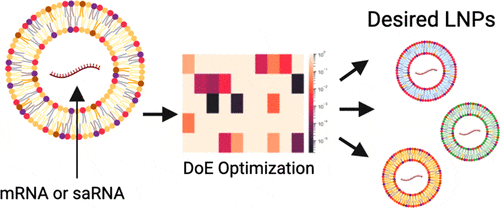
Specialized software, such as Design-Expert®, JMP®, or Minitab®, facilitates the design and analysis of experiments, making the implementation of DOE approaches more accessible and effective.
LNP Formulation Screening Using Artificial Intelligence (AI)
Given the vast amount of data involved in LNP formulation, AI is increasingly being investigated as a tool to predict the most successful formulations. AI has already proven its utility in the rational design of ionizable lipids, as demonstrated in recent research (Reference). Its trajectory suggests significant potential for broader applications in the LNP field.
However, when it comes to the overall formulation process, significant limitations remain. As highlighted by Prof. Christine Allen from the University of Toronto during The Nanomedicine and Drug Delivery Symposium in Orlando, FL (2024), several challenges impede AI’s application in LNP screening:
- Lack of Uniformity: Variations in LNP preparation methods and inconsistencies in reporting experimental conditions make standardization difficult.
- Incomplete Data: Many scientific publications lack comprehensive reporting of formulation conditions, hindering the development of robust datasets for AI models.
- High Data Requirements: The sheer volume of data needed to train effective AI models poses a barrier, especially when considering the diverse parameters influencing LNP performance.
Despite these challenges, AI remains a promising tool for future advancements in LNP screening. By addressing the current limitations through improved reporting standards, data collection, and collaborative efforts, AI could complement traditional DOE approaches, paving the way for more efficient and predictive LNP formulation strategies.
A practical approach to LNP screening
The importance of the right choice of LNP formulation method
The formulation process and associated parameters significantly impact nanoparticle CQAs, making it critical to address these factors early in the development process. Neglecting to do so can lead to substantial disruptions during scale-up, compromising nanoparticle quality and therapeutic outcomes.
Screening Requirements
Screening processes typically follow a two-step approach:
In Vitro Testing:
This step evaluates key hits from a broader pool of formulations. It involves physicochemical characterization and initial cell-based assays to identify promising candidates.
In Vivo Testing:
Once the in vitro hits are identified, they undergo more detailed in vivo screening to validate efficacy and safety. Due to the cost and ethical considerations of animal testing, this step is minimized and focused on the most promising LNP candidates to eventually determine the lead LNP formulation.
During in vitro testing, the goal is to characterize a wide range of LNP formulations using minimal volumes. Typically, a few hundred microliters (100s of µL) suffice for essential tests like dynamic light scattering (DLS) for size, PDI, and zeta potential; encapsulation efficiency; and initial cell-based assays. For in vivo testing, formulation volumes may increase to the milliliter scale, depending on the number of injections and boosters required.
High throughput in vivo screening
Recently, several universities and companies have developed novel high-throughput approaches for directly testing RNA-LNP formulations in vivo, bypassing traditional in vitro testing, which can sometimes yield unreliable results. These innovative methods involve the simultaneous injection of multiple RNA-LNP formulations, each uniquely labeled with a DNA barcode attached to the nucleic acids. These barcodes enable precise tracking and evaluation of the biodistribution and efficacy of each RNA-LNP formulation independently, providing valuable insights into their performance in a biologically relevant environment.
Why using microfluidics for LNP formulation screening is key ?
Whichever the approach used (in vitro then vivo, or high throughput in vivo screening) , microfluidics formulation of LNP emerges as the most suitable approach for low-volume, high-precision LNP production requirements for screening. Unlike manual mixing, which suffers from poor reproducibility and limited control, microfluidics allows for precise regulation of formulation parameters, ensuring consistent and reproducible CQAs even at small scales. Furthermore, all the other methods cannot accommodate µL-scale formulations making them impractical due to their high operational costs. These advantages make microfluidics the optimal choice for the iterative and resource-intensive nature of LNP screening processes.
A practical example of RNA-LNP screening process with microfluidics using TAMARA
In collaboration with the ART-ARNm INSERM laboratory, a leader in RNA-LNP formulation in France, we conducted a basic LNP screening study using the TAMARA microfluidic platform. The goal was to evaluate the influence of formulation process parameters on a specific LNP composition using internally developed lipids and to compare TAMARA’s performance to other microfluidic methods.
Volume & reproducibility:
Initial tests asses the effect of formulation volume on LNP size & PDI to identify the ideal formulation volume.
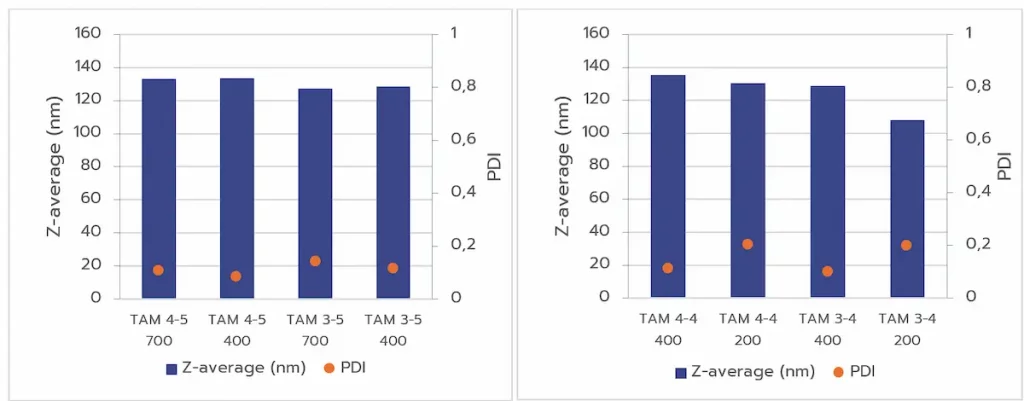
Formulations at 400 µL and 700 µL produced consistent results, with LNP size in the perfect range, while 200 µL volumes exhibited higher variability and larger PDI. Based on these results, a minimum volume of 400 µL was established for subsequent experiments.
Encapsulation Efficiency and Yield:
In a second step, encapsulation efficiency was tested at different volumes.
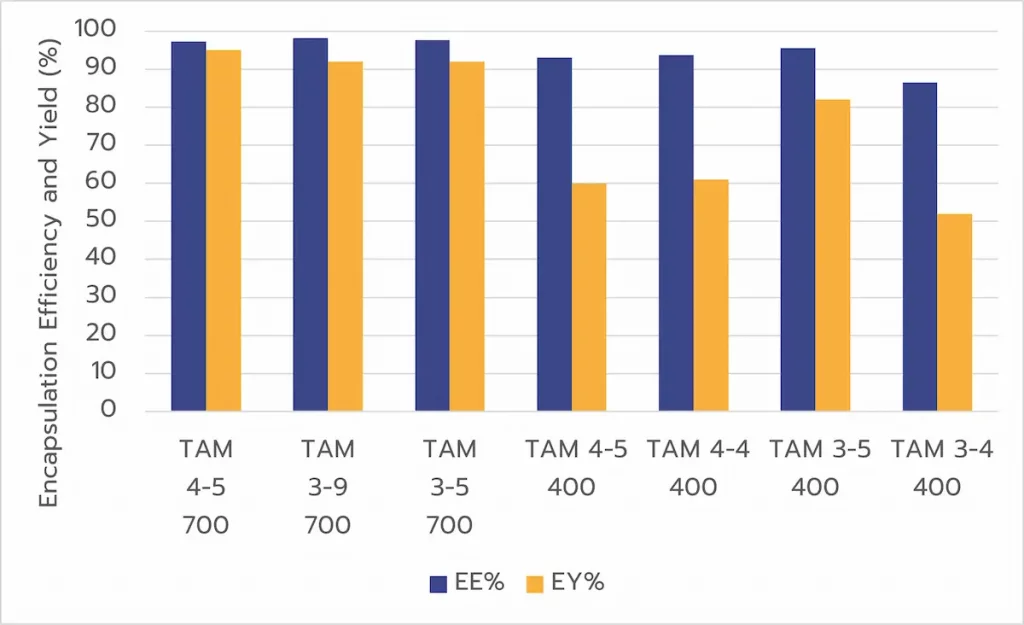
TAMARA achieved encapsulation efficiencies exceeding 90% even at low volumes. However, encapsulation yield (EY%) was slightly lower at smaller scales, highlighting opportunities for further optimization.
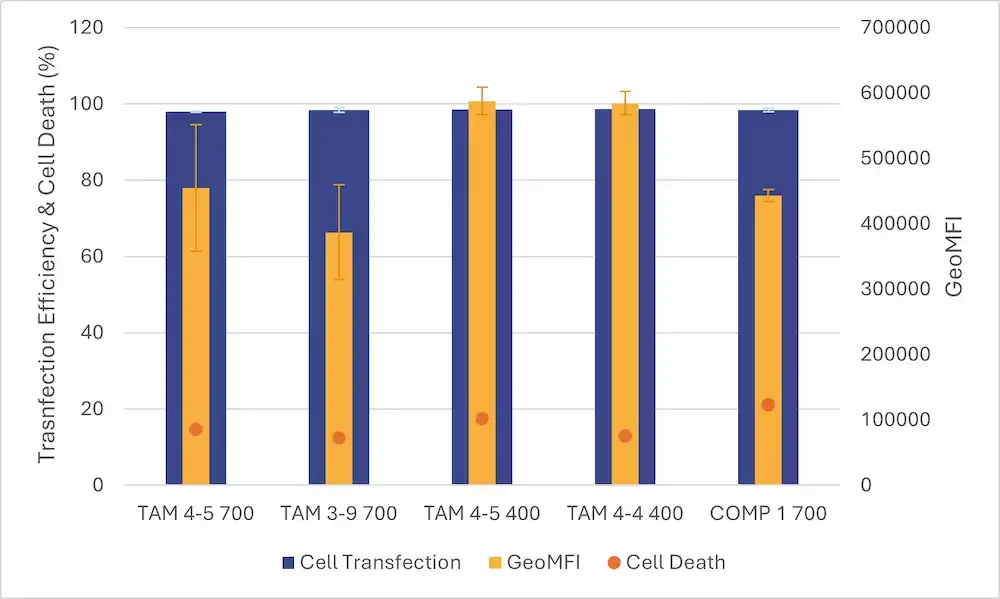
In parallel, the TAMARA was also compared to another standard microfluidic approach (toroidal mixer such as NanoAssemblr Ignite by Precision Nanosystems) previously optimized through a DOE at identical volumes.
When comparing the 2 approaches, TAMARA delivered similar or better encapsulation efficiencies (+5%) and superior RNA recovery (+20%), resulting in significantly improved overall outcomes.
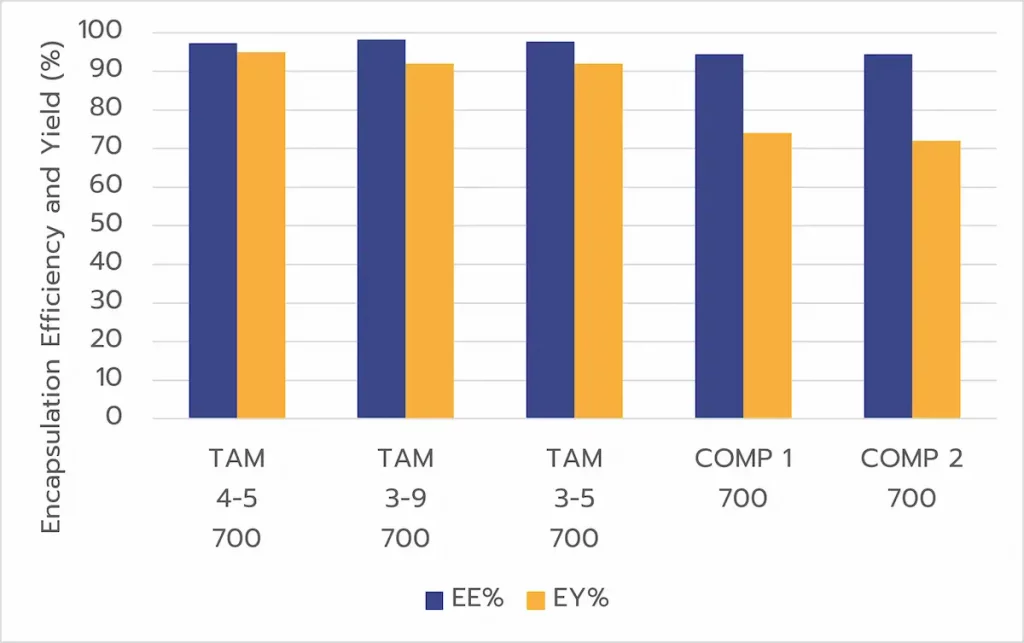
Cell-based testing
Finally, a series of tests was carried out on cells to identify the main hits.
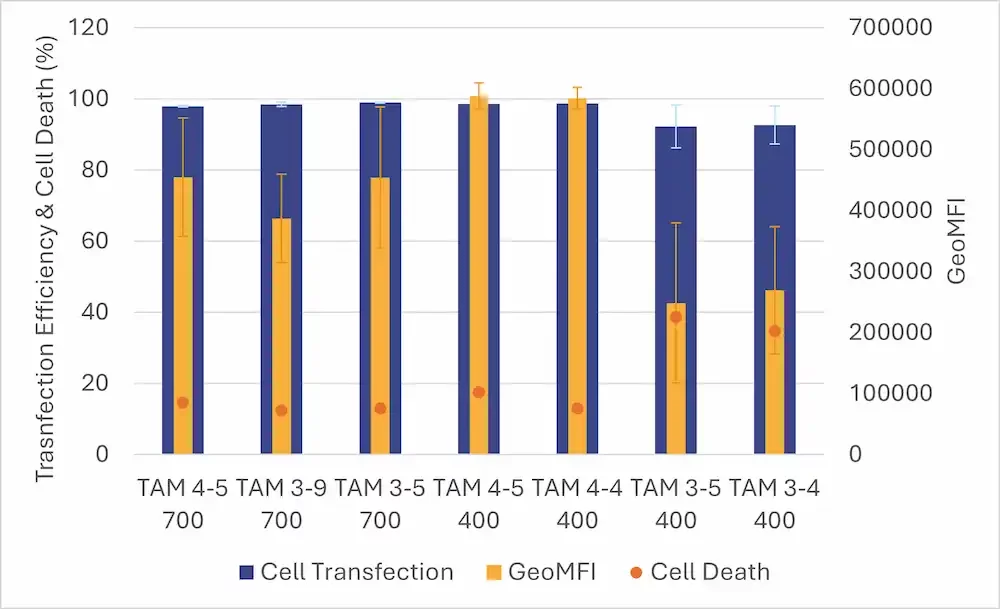
Cell assays revealed consistent transfection efficiencies (>90%) across the different LNP formulations produced with TAMARA. By modifying formulation parameters, significant differences in protein expression (GeoMFI) and viability were observed.
Compared to other microfluidic approaches optimized via DOE, TAMARA demonstrated equal or better protein expression outcomes, similar transfection efficiency and superior cell viability, even at lower formulation volumes.
This study underscores the deep impact of formulation processes and parameters on LNP CQAs. Even minor variations in a single CPP can significantly influence the final therapeutic outcome. As demonstrated, microfluidics—and TAMARA in particular—offers a robust, reproducible platform for efficient LNP screening, enabling precise tuning of CPPs and facilitating the identification of optimal formulations while minimizing RNA & lipid consumption.
To learn more, check out our thorough RNA-LNP Formulation screening with TAMARA application note.
Conclusion on LNP formulation screening
LNP formulation screening is a complex but indispensable step in the development of RNA-based therapeutics. By understanding the interplay between critical process parameters (CPPs) critical quality attributes (CQAs) and Quality Target Product Profile (QTPP) drug developers can make informed decisions to optimize formulations for safety, efficacy, and reproducibility. However, the number of tests to carry out to evaluate the whole space is massive, with the cost of RNA & lipids hindering it.
Advanced tools & methodologies like Design of Experiments (DOE) and Artificial Intelligence (AI) allow for the minimization of the number of formulation tested, yet they should be combined with state-of-the-art formulation methods such as microfluidics, to significantly streamline this process and minimize RNA consumption, enabling precise and efficient exploration of vast experimental spaces while keeping cost as low as possible.
As the field advances, however, the need for scalable solutions that seamlessly bridge low-volume screening and large-scale formulation production is becoming increasingly apparent. These solutions are essential to maintain QTPP consistency across scales, paving the way for efficient and reproducible drug development. At Inside Therapeutics, we are addressing this challenge with our forthcoming NanoPulse technology, which promises to deliver an innovative solution. Stay tuned for more details in an upcoming article!

Looking to start or improve your LNP formulation screening?
Reach out to us to learn how we can help!
References
[1] Li B, Manan RS, Liang SQ, et al. Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing. Nat Biotechnol. 2023;41(10):1410-1415. doi:10.1038/s41587-023-01679-x
[2] Schoenmaker L, Witzigmann D, Kulkarni JA, et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int J Pharm. 2021;601:120586. doi:10.1016/j.ijpharm.2021.120586





